library(Seurat)
library(SeuratData)
library(BayesSpace)
library(ggplot2)
source("Cours/FunctionsAux.R")TP - Partie 3
Pour aller plus loin
1 Spatial transcriptomic (ST)
1.1 Données de l’exemple
Description:
Données de transcirptomique spatiale du cerveau de la souris pour la région antérieure
Expression de \(G=31053\) gènes pour \(C=2696\) spots
InstallData("stxBrain")Warning: The following packages are already installed and will not be
reinstalled: stxBrainbrain <- LoadData("stxBrain", type = "anterior1")Validating object structureUpdating object slotsEnsuring keys are in the proper structure
Ensuring keys are in the proper structureEnsuring feature names don't have underscores or pipesUpdating slots in SpatialUpdating slots in anterior1Validating object structure for Assay5 'Spatial'Validating object structure for VisiumV2 'anterior1'Object representation is consistent with the most current Seurat versionbrainAn object of class Seurat
31053 features across 2696 samples within 1 assay
Active assay: Spatial (31053 features, 0 variable features)
1 layer present: counts
1 spatial field of view present: anterior1brain@assays$Spatial$counts[1:6,1:10]6 x 10 sparse Matrix of class "dgCMatrix" [[ suppressing 10 column names 'AAACAAGTATCTCCCA-1', 'AAACACCAATAACTGC-1', 'AAACAGAGCGACTCCT-1' ... ]]
Xkr4 . . . . . . . . . .
Gm1992 . . . . . . . . . .
Gm37381 . . . . . . . . . .
Rp1 . . . . . . . . . .
Sox17 . 1 . . . 2 . . 1 .
Gm37323 . . . . . . . . . .brain@images$anterior1Spatial coordinates for 2696 cells
Default segmentation boundary: centroids
Associated assay: Spatial
Key: slice1_ SpatialFeaturePlot(brain, features = "nCount_Spatial") +
theme(legend.position = "right")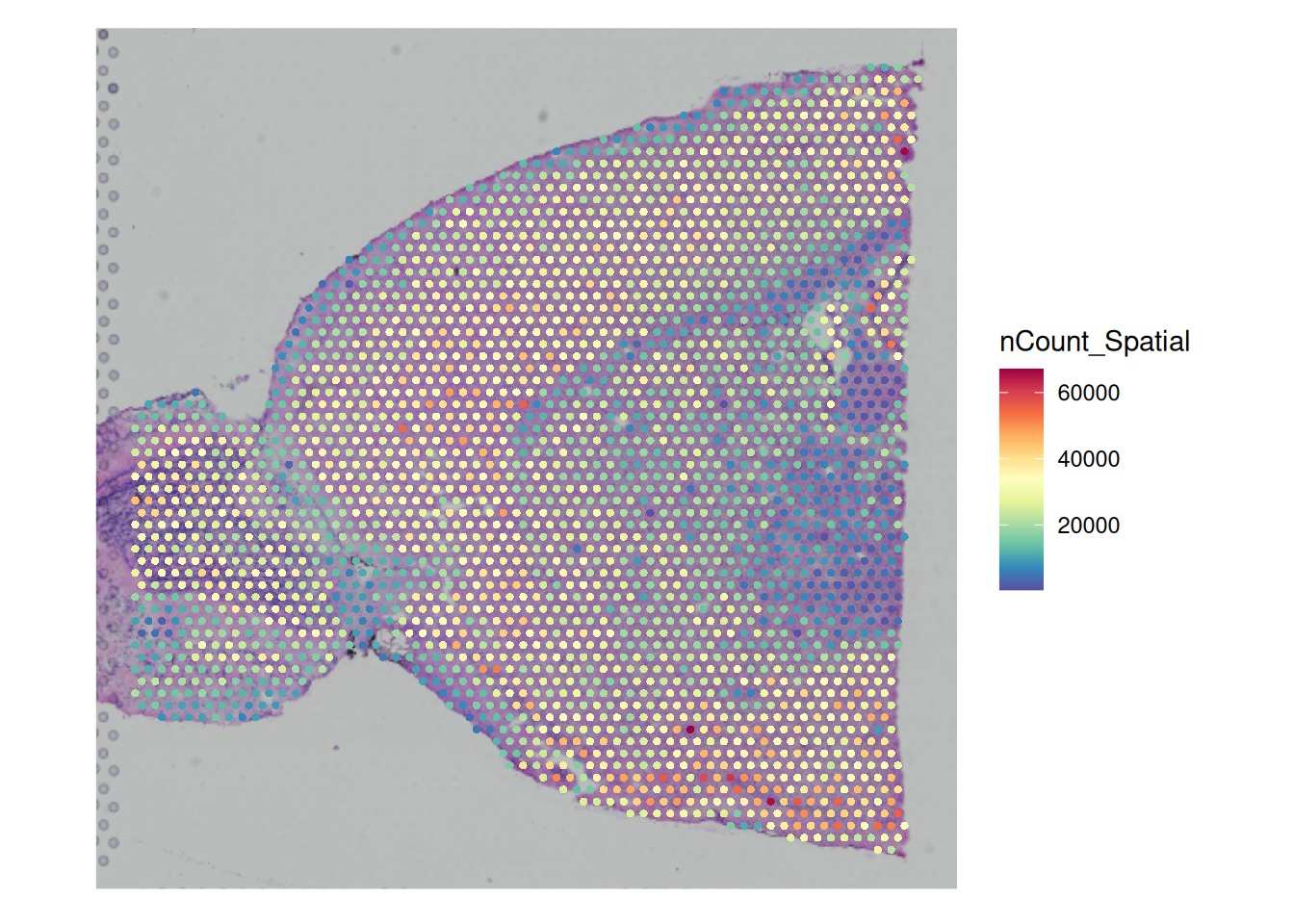
1.2 Normalisation avec SCTransform
- Comprend les étapes de normalisation, calcul des HVG et scale data
brain <- SCTransform(brain, assay = "Spatial", verbose = FALSE)
brain@assays$SCTSCTAssay data with 17668 features for 2696 cells, and 1 SCTModel(s)
Top 10 variable features:
Ttr, Hbb-bs, Mbp, Plp1, Hba-a1, Ptgds, Penk, S100a5, Hba-a2, Fabp7 plot1<-VariableFeaturePlot(brain)
top10<-head(VariableFeatures(brain), 10)
plot2 <- LabelPoints(plot = plot1, points = top10, repel = TRUE)When using repel, set xnudge and ynudge to 0 for optimal resultsplot2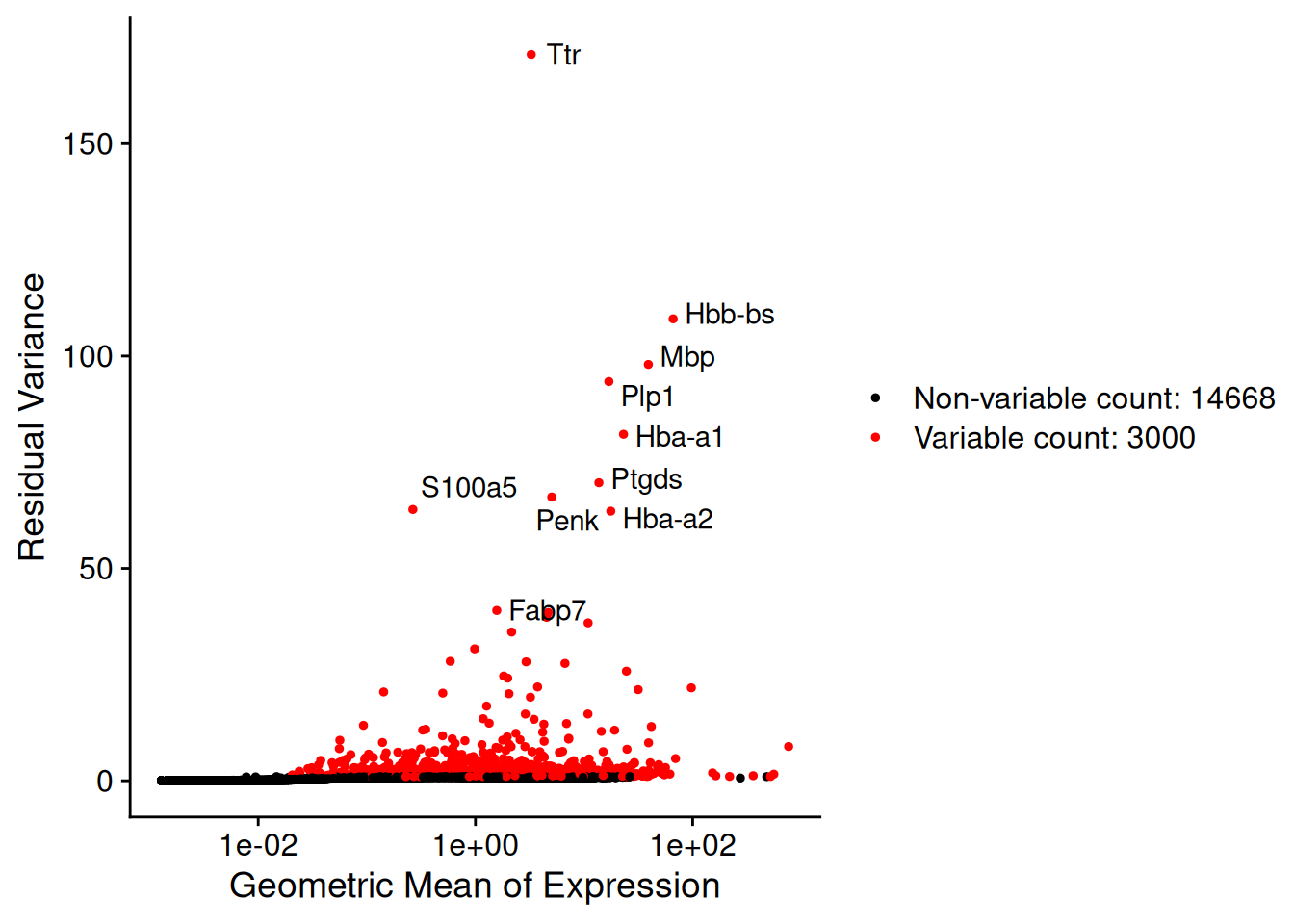
SpatialFeaturePlot(brain, features = c("Ttr","Hbb-bs"))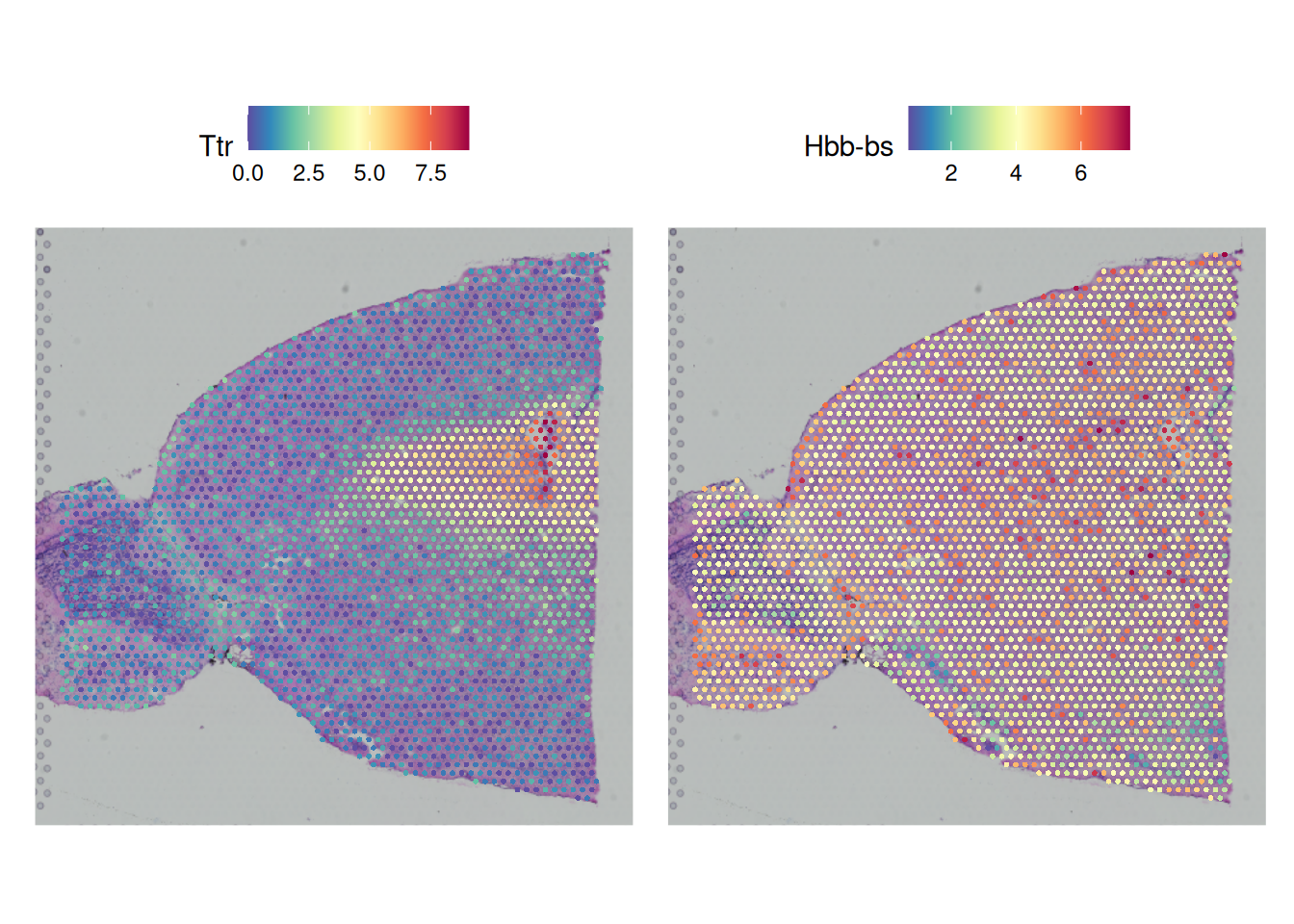
1.3 Réduction de dimension
brain <- RunPCA(brain, assay = "SCT", verbose = FALSE)
brain <- FindNeighbors(brain, reduction = "pca", dims = 1:30)Computing nearest neighbor graphComputing SNNbrain <- RunUMAP(brain, reduction = "pca", dims = 1:30)Warning: The default method for RunUMAP has changed from calling Python UMAP via reticulate to the R-native UWOT using the cosine metric
To use Python UMAP via reticulate, set umap.method to 'umap-learn' and metric to 'correlation'
This message will be shown once per session13:14:31 UMAP embedding parameters a = 0.9922 b = 1.11213:14:31 Read 2696 rows and found 30 numeric columns13:14:31 Using Annoy for neighbor search, n_neighbors = 3013:14:31 Building Annoy index with metric = cosine, n_trees = 500% 10 20 30 40 50 60 70 80 90 100%[----|----|----|----|----|----|----|----|----|----|**************************************************|
13:14:31 Writing NN index file to temp file /tmp/RtmpZRDtDb/file5ed660a942a6
13:14:31 Searching Annoy index using 1 thread, search_k = 3000
13:14:32 Annoy recall = 100%
13:14:33 Commencing smooth kNN distance calibration using 1 thread with target n_neighbors = 30
13:14:36 Initializing from normalized Laplacian + noise (using RSpectra)
13:14:36 Commencing optimization for 500 epochs, with 105150 positive edges
13:14:36 Using rng type: pcg
13:14:41 Optimization finishedDimPlot(brain, reduction = "umap")+NoLegend()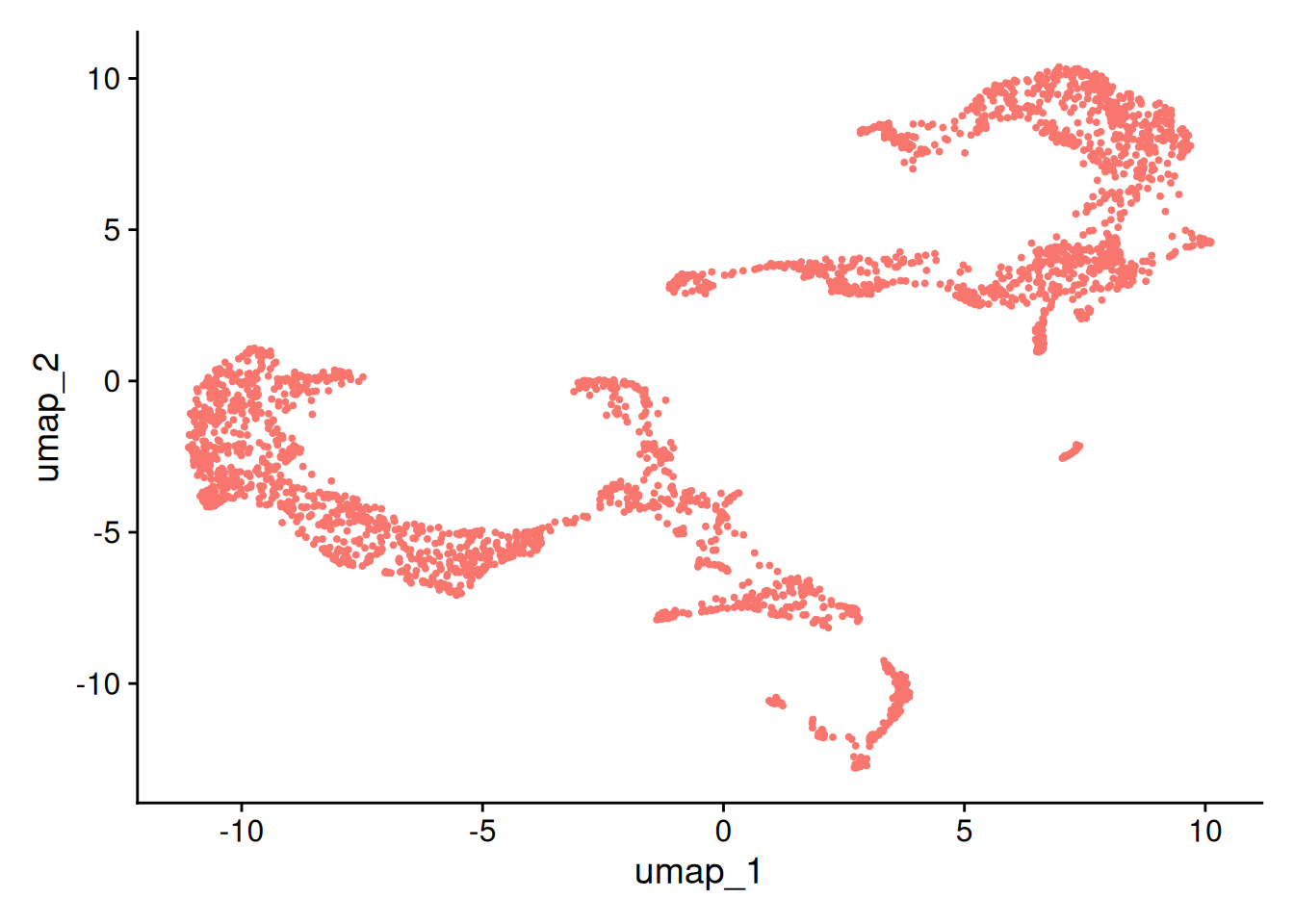
1.4 Clustering
- Exemple avec BayesSpace
diet.seurat = Seurat::DietSeurat(brain,
graphs = "SCT_nn",
dimreducs = c("pca"))
sce_brain = as.SingleCellExperiment(diet.seurat, assay = "SCT")
colData(sce_brain) = cbind(colData(sce_brain),
brain@images$anterior1$centroids@coords)
sce_brain$array_col<-sce_brain$y
sce_brain$array_row<-sce_brain$x
sce_brain = spatialPreprocess(sce_brain,
platform = "Visium",
skip.PCA = T,
log.normalize = log.normalize)
sce_brain <- spatialCluster(sce_brain,
nrep = 1000,
q= 8,
burn.in = 10,
model = "t",
gamma = 2)
brain@meta.data[["BayesSpace"]] <- as.factor(sce_brain$spatial.cluster)EffPlot(brain,clustname="BayesSpace")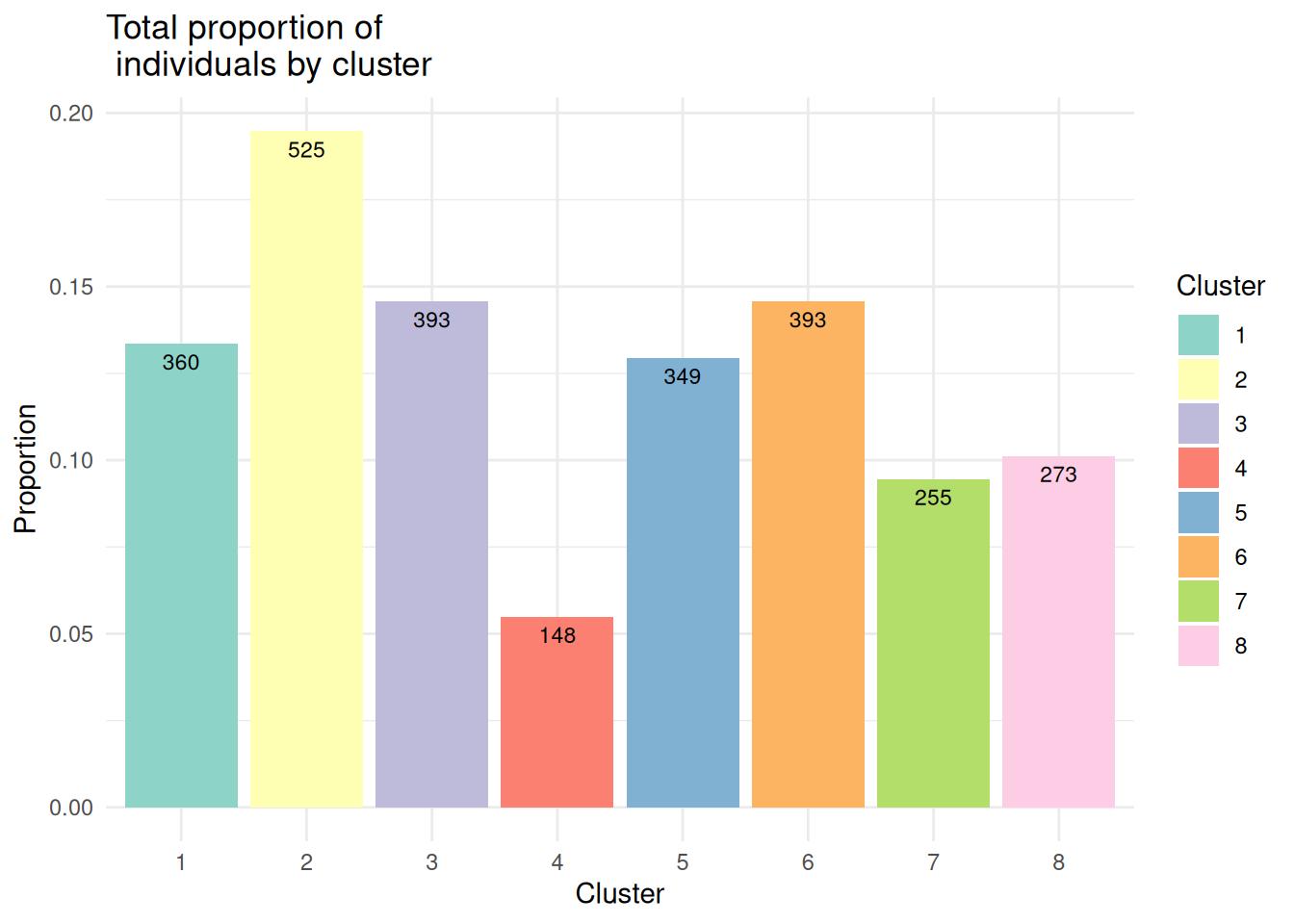
p1<-SpatialDimPlot(brain, "BayesSpace",label=TRUE,label.size=2)Scale for fill is already present.
Adding another scale for fill, which will replace the existing scale.p2<-DimPlot(brain, reduction = "umap",group.by="BayesSpace",label=TRUE)
p1+p2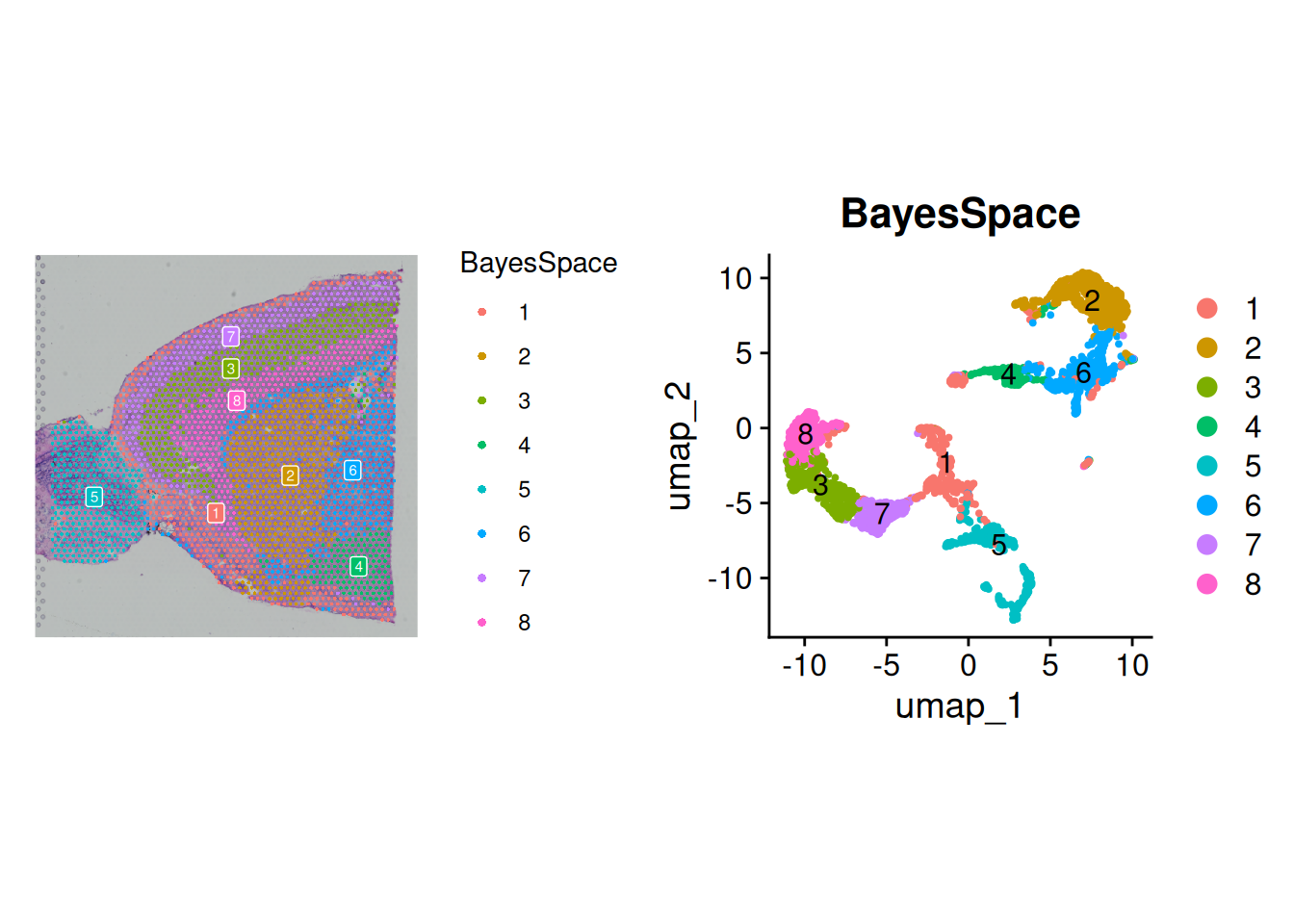
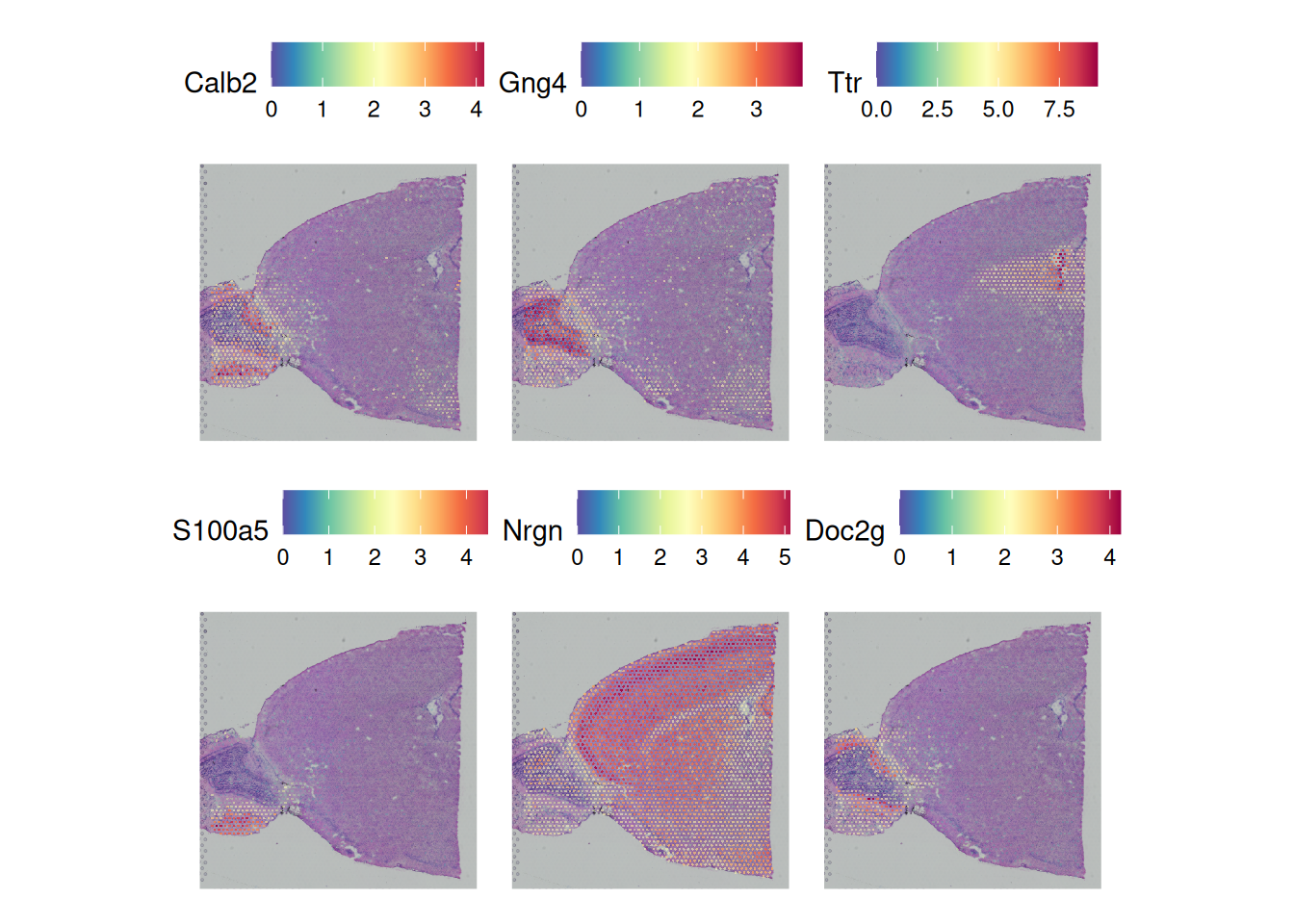
1.5 Spatial Variable Features
brain <- FindSpatiallyVariableFeatures(brain,
assay = "SCT",
features=VariableFeatures(brain)[1:1000],
selection.method = "moransi")top.features = rownames(
dplyr::slice_min(
brain[["SCT"]]@meta.features,
moransi.spatially.variable.rank,
n = 6
)
)
SpatialFeaturePlot(brain, features = top.features, ncol = 3, alpha = c(0.1, 1))